The spread of a plant pathogen along the former trade routes of the Silk Road
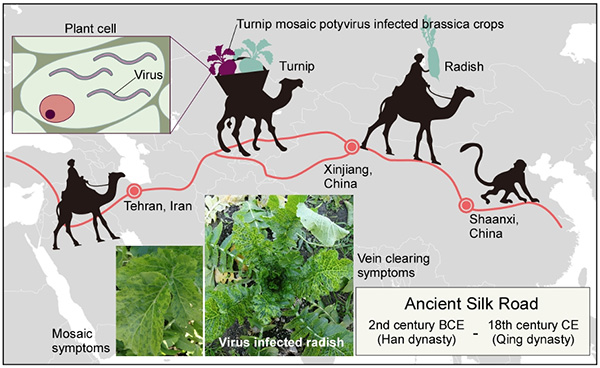 For nearly two thousand years, the Silk Road was an important network of trade routes connecting Europe to East Asia via the Middle East. The Silk Road allowed the exchange of culture, religion, livestock, and crops. It also provided a pathway for the spread of plant pathogens, as shown in a genomic analysis of turnip mosaic potyvirus published in Proceedings of the National Academy of Sciences of the USA.
For nearly two thousand years, the Silk Road was an important network of trade routes connecting Europe to East Asia via the Middle East. The Silk Road allowed the exchange of culture, religion, livestock, and crops. It also provided a pathway for the spread of plant pathogens, as shown in a genomic analysis of turnip mosaic potyvirus published in Proceedings of the National Academy of Sciences of the USA.
A research group from Saga University, working with international colleagues, has reconstructed the historical spread of turnip mosaic potyvirus across Eurasia. This virus is usually transmitted by aphids and is a significant pathogen of brassica vegetables, a group of crops that include turnip, cauliflower, and broccoli. The authors found that the migration of the virus traced some of the major trade arteries of the Silk Road.
This study also presents the most comprehensive reconstruction of the evolutionary and epidemiological history of a major plant pathogen of agricultural crops. By analyzing more than 500 genomes of the virus, sampled from throughout Eurasia, the study is the first to describe the spatial and temporal patterns of migration for a plant pathogen on such a scale. Much of the collection was done on foot during expeditions led by Professor Kazusato Ohshima. He said: “We spent more than a quarter of a century collecting samples of this virus, but our sampling is still incomplete”. His student Shusuke Kawakubo, the first author of the study, said: “It is impressive how the virus travelled across the world, and how its evolution has been influenced by cultural activities”.
By using statistical methods to analyse the genome data, Professor Ohshima and colleagues found that the virus emerged in the Middle East and began its spread across Eurasia in the 17th century. It then split into different lineages according to the host plants in different parts of the world. Professor Simon Ho (University of Sydney), who collaborated on the project, said: “Computer analyses of virus genomes can tell us so much about the history of important pathogens, especially when have such a detailed sample of the virus from throughout its range”.
Human viruses typically spread more quickly than plant pathogens. There is no more convincing example of this than SARS-Cov-2, the causative agent of COVID-19, which has left no corner of the world untouched. Plant viruses have migrated more slowly, as shown in this study of turnip mosaic potyvirus, even though the genomes of human and plant viruses evolve at similar rates. Plant protection is the front line of protection against the epidemic spread of plant pathogens. It is important to keep those notorious invaders under strict surveillance, particularly because human-mediated transmission should be controllable.
Reference
Kawakubo S, Gao F, Li S, Tan Z, Huang Y-K, Adkar-Purushothama CR, Gurikar C, Maneechoat P, Chiemsombat P, Aye SS, Furuya N, Shevchenko O, Špak J, Škorić D, Ho SYW, Ohshima K. 2021. Genomic analysis of the brassica pathogen turnip mosaic potyvirus reveals its spread along the former trade routes of the Silk Road. Proceedings of the National Academy of Sciences of the USA 118, e2021221118.
